step_downsample() creates a specification of a recipe step that will
remove rows of a data set to make the occurrence of levels in a specific
factor level equal.
Usage
step_downsample(
recipe,
...,
under_ratio = 1,
ratio = deprecated(),
role = NA,
trained = FALSE,
column = NULL,
target = NA,
skip = TRUE,
seed = sample.int(10^5, 1),
id = rand_id("downsample")
)Arguments
- recipe
A recipe object. The step will be added to the sequence of operations for this recipe.
- ...
One or more selector functions to choose which variable is used to sample the data. See recipes::selections for more details. The selection should result in single factor variable. For the
tidymethod, these are not currently used.- under_ratio
A numeric value for the ratio of the majority-to-minority frequencies. The default value (1) means that all other levels are sampled down to have the same frequency as the least occurring level. A value of 2 would mean that the majority levels will have (at most) (approximately) twice as many rows than the minority level.
- ratio
Deprecated argument; same as
under_ratio- role
Not used by this step since no new variables are created.
- trained
A logical to indicate if the quantities for preprocessing have been estimated.
- column
A character string of the variable name that will be populated (eventually) by the
...selectors.- target
An integer that will be used to subsample. This should not be set by the user and will be populated by
prep.- skip
A logical. Should the step be skipped when the recipe is baked by
bake()? While all operations are baked whenprep()is run, some operations may not be able to be conducted on new data (e.g. processing the outcome variable(s)). Care should be taken when usingskip = TRUEas it may affect the computations for subsequent operations.- seed
An integer that will be used as the seed when downsampling.
- id
A character string that is unique to this step to identify it.
Value
An updated version of recipe with the new step
added to the sequence of existing steps (if any). For the
tidy method, a tibble with columns terms which is
the variable used to sample.
Details
Down-sampling is intended to be performed on the training set
alone. For this reason, the default is skip = TRUE.
If there are missing values in the factor variable that is used to define the sampling, missing data are selected at random in the same way that the other factor levels are sampled. Missing values are not used to determine the amount of data in the minority level
For any data with factor levels occurring with the same frequency as the minority level, all data will be retained.
All columns in the data are sampled and returned by recipes::juice()
and recipes::bake().
Keep in mind that the location of down-sampling in the step may have effects. For example, if centering and scaling, it is not clear whether those operations should be conducted before or after rows are removed.
Tidying
When you tidy() this step, a tibble is retruned with
columns terms and id:
- terms
character, the selectors or variables selected
- id
character, id of this step
Tuning Parameters
This step has 1 tuning parameters:
under_ratio: Under-Sampling Ratio (type: double, default: 1)
Case weights
This step performs an unsupervised operation that can utilize case weights.
To use them, see the documentation in recipes::case_weights and the examples on
tidymodels.org.
See also
Other Steps for under-sampling:
step_nearmiss(),
step_tomek()
Examples
library(recipes)
library(modeldata)
data(hpc_data)
hpc_data0 <- hpc_data |>
select(-protocol, -day)
orig <- count(hpc_data0, class, name = "orig")
orig
#> # A tibble: 4 × 2
#> class orig
#> <fct> <int>
#> 1 VF 2211
#> 2 F 1347
#> 3 M 514
#> 4 L 259
up_rec <- recipe(class ~ ., data = hpc_data0) |>
# Bring the majority levels down to about 1000 each
# 1000/259 is approx 3.862
step_downsample(class, under_ratio = 3.862) |>
prep()
training <- up_rec |>
bake(new_data = NULL) |>
count(class, name = "training")
training
#> # A tibble: 4 × 2
#> class training
#> <fct> <int>
#> 1 VF 1000
#> 2 F 1000
#> 3 M 514
#> 4 L 259
# Since `skip` defaults to TRUE, baking the step has no effect
baked <- up_rec |>
bake(new_data = hpc_data0) |>
count(class, name = "baked")
baked
#> # A tibble: 4 × 2
#> class baked
#> <fct> <int>
#> 1 VF 2211
#> 2 F 1347
#> 3 M 514
#> 4 L 259
# Note that if the original data contained more rows than the
# target n (= ratio * majority_n), the data are left alone:
orig |>
left_join(training, by = "class") |>
left_join(baked, by = "class")
#> # A tibble: 4 × 4
#> class orig training baked
#> <fct> <int> <int> <int>
#> 1 VF 2211 1000 2211
#> 2 F 1347 1000 1347
#> 3 M 514 514 514
#> 4 L 259 259 259
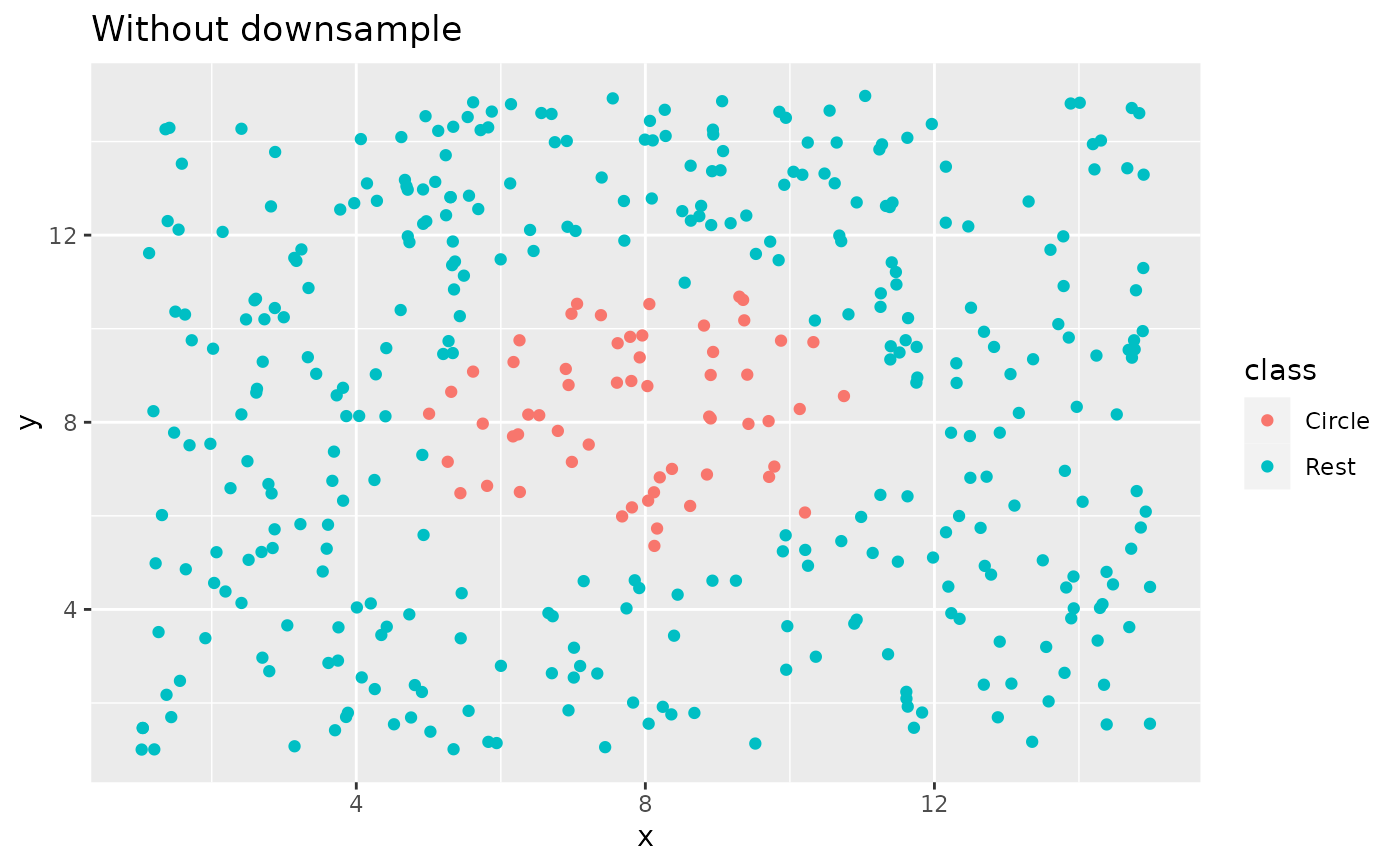
library(ggplot2)
ggplot(circle_example, aes(x, y, color = class)) +
geom_point() +
labs(title = "Without downsample")
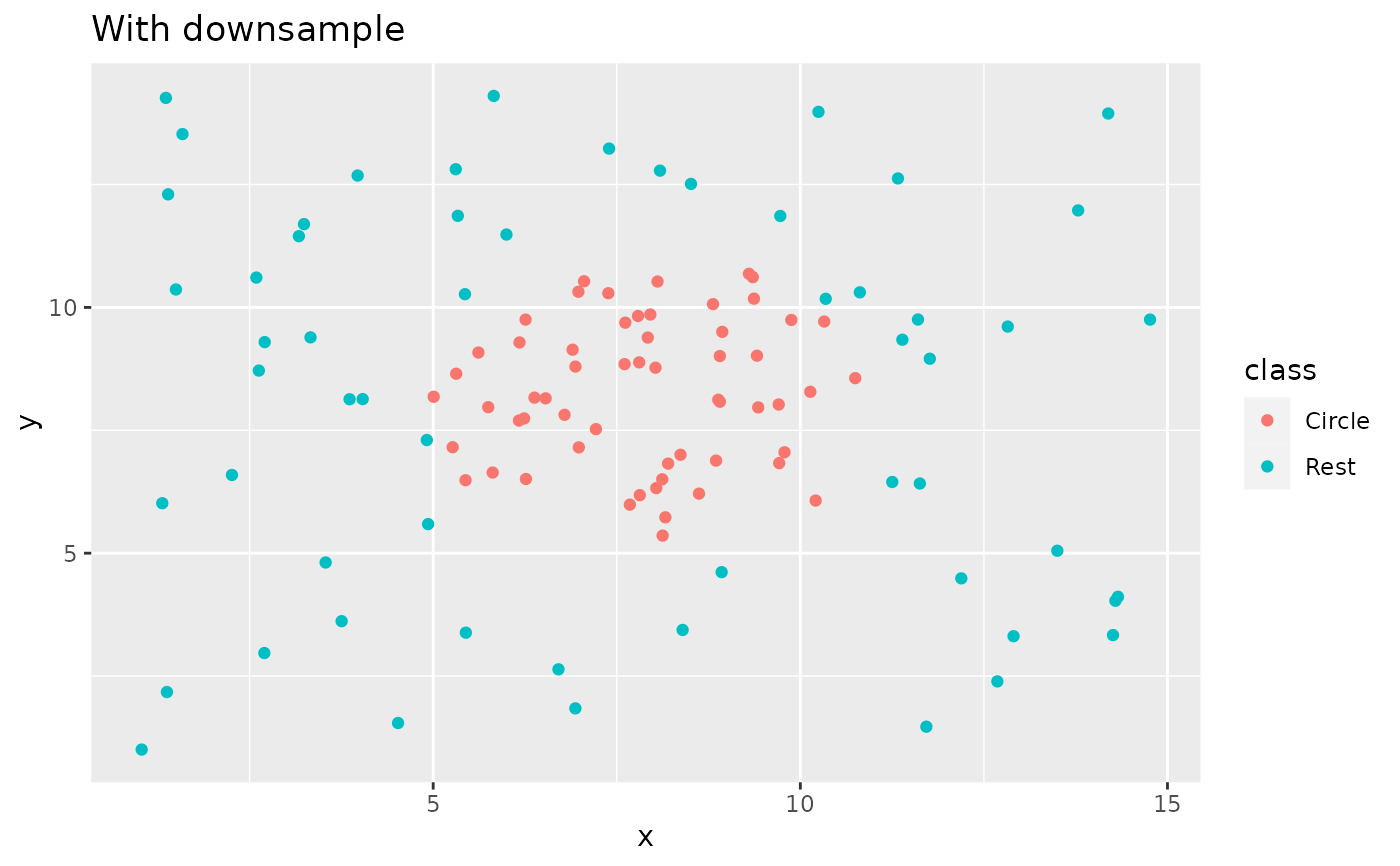
 recipe(class ~ x + y, data = circle_example) |>
step_downsample(class) |>
prep() |>
bake(new_data = NULL) |>
ggplot(aes(x, y, color = class)) +
geom_point() +
labs(title = "With downsample")
recipe(class ~ x + y, data = circle_example) |>
step_downsample(class) |>
prep() |>
bake(new_data = NULL) |>
ggplot(aes(x, y, color = class)) +
geom_point() +
labs(title = "With downsample")