step_tomek() creates a specification of a recipe step that removes
majority class instances of tomek links.
Usage
step_tomek(
recipe,
...,
role = NA,
trained = FALSE,
column = NULL,
skip = TRUE,
seed = sample.int(10^5, 1),
id = rand_id("tomek")
)Arguments
- recipe
A recipe object. The step will be added to the sequence of operations for this recipe.
- ...
One or more selector functions to choose which variable is used to sample the data. See recipes::selections for more details. The selection should result in single factor variable. For the
tidymethod, these are not currently used.- role
Not used by this step since no new variables are created.
- trained
A logical to indicate if the quantities for preprocessing have been estimated.
- column
A character string of the variable name that will be populated (eventually) by the
...selectors.- skip
A logical. Should the step be skipped when the recipe is baked by
bake()? While all operations are baked whenprep()is run, some operations may not be able to be conducted on new data (e.g. processing the outcome variable(s)). Care should be taken when usingskip = TRUEas it may affect the computations for subsequent operations.- seed
An integer that will be used as the seed when applied.
- id
A character string that is unique to this step to identify it.
Value
An updated version of recipe with the new step
added to the sequence of existing steps (if any). For the
tidy method, a tibble with columns terms which is
the variable used to sample.
Details
The factor variable used to balance around must only have 2 levels. All other variables must be numerics with no missing data.
A tomek link is defined as a pair of points from different classes and are each others nearest neighbors.
All columns in the data are sampled and returned by recipes::juice()
and recipes::bake().
When used in modeling, users should strongly consider using the
option skip = TRUE so that the extra sampling is not
conducted outside of the training set.
Tidying
When you tidy() this step, a tibble is retruned with
columns terms and id:
- terms
character, the selectors or variables selected
- id
character, id of this step
See also
tomek() for direct implementation
Other Steps for under-sampling:
step_downsample(),
step_nearmiss()
Examples
library(recipes)
library(modeldata)
data(hpc_data)
hpc_data0 <- hpc_data %>%
select(-protocol, -day)
orig <- count(hpc_data0, class, name = "orig")
orig
#> # A tibble: 4 × 2
#> class orig
#> <fct> <int>
#> 1 VF 2211
#> 2 F 1347
#> 3 M 514
#> 4 L 259
up_rec <- recipe(class ~ ., data = hpc_data0) %>%
step_tomek(class) %>%
prep()
training <- up_rec %>%
bake(new_data = NULL) %>%
count(class, name = "training")
training
#> # A tibble: 4 × 2
#> class training
#> <fct> <int>
#> 1 VF 1911
#> 2 F 1011
#> 3 M 379
#> 4 L 210
# Since `skip` defaults to TRUE, baking the step has no effect
baked <- up_rec %>%
bake(new_data = hpc_data0) %>%
count(class, name = "baked")
baked
#> # A tibble: 4 × 2
#> class baked
#> <fct> <int>
#> 1 VF 2211
#> 2 F 1347
#> 3 M 514
#> 4 L 259
orig %>%
left_join(training, by = "class") %>%
left_join(baked, by = "class")
#> # A tibble: 4 × 4
#> class orig training baked
#> <fct> <int> <int> <int>
#> 1 VF 2211 1911 2211
#> 2 F 1347 1011 1347
#> 3 M 514 379 514
#> 4 L 259 210 259
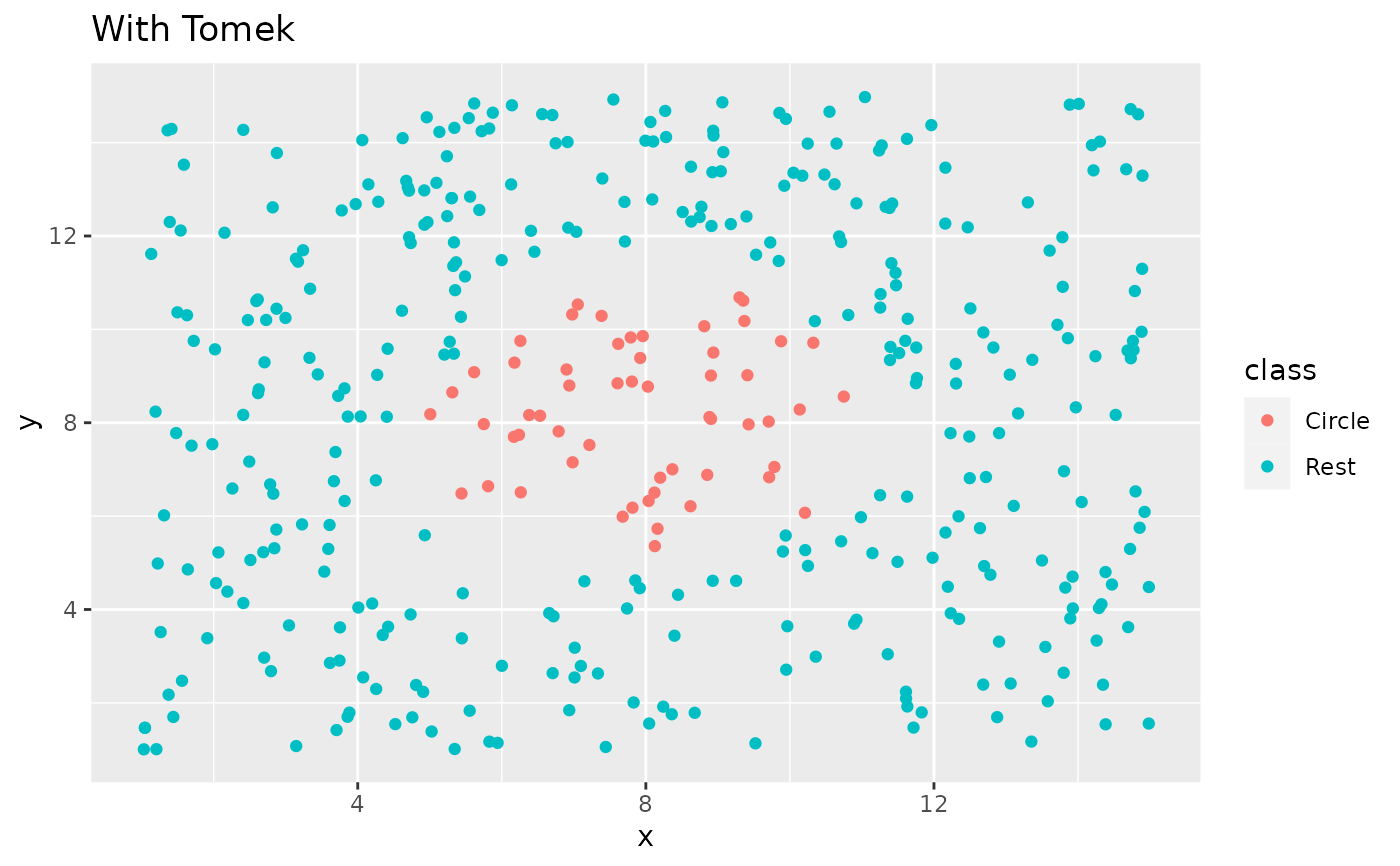
library(ggplot2)
ggplot(circle_example, aes(x, y, color = class)) +
geom_point() +
labs(title = "Without Tomek") +
xlim(c(1, 15)) +
ylim(c(1, 15))
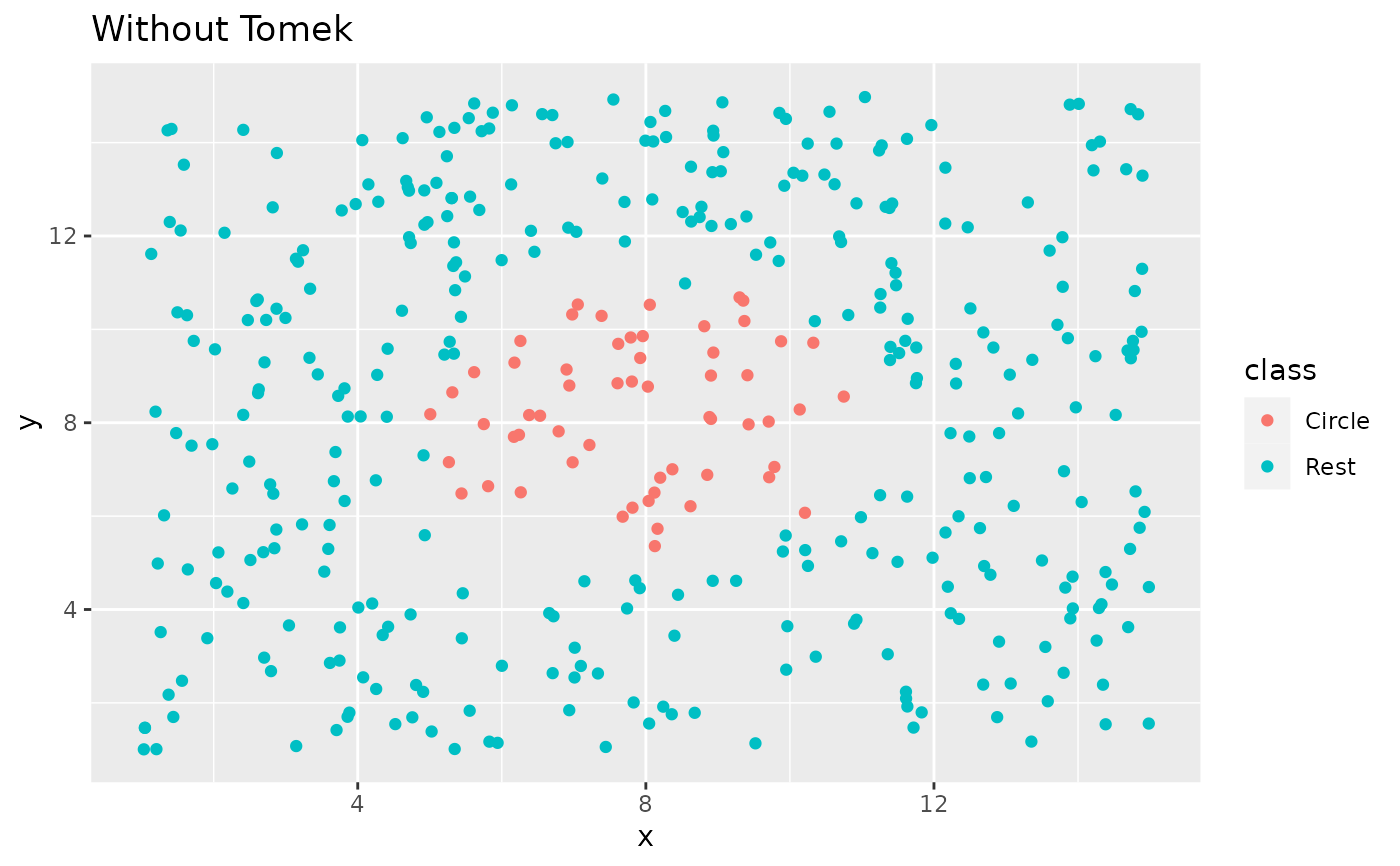 recipe(class ~ x + y, data = circle_example) %>%
step_tomek(class) %>%
prep() %>%
bake(new_data = NULL) %>%
ggplot(aes(x, y, color = class)) +
geom_point() +
labs(title = "With Tomek") +
xlim(c(1, 15)) +
ylim(c(1, 15))
recipe(class ~ x + y, data = circle_example) %>%
step_tomek(class) %>%
prep() %>%
bake(new_data = NULL) %>%
ggplot(aes(x, y, color = class)) +
geom_point() +
labs(title = "With Tomek") +
xlim(c(1, 15)) +
ylim(c(1, 15))